Serial analysis of gene expression (SAGE) is a technique used to provide a library of the mRNA sequences (transcripts) of the sample in the form of small tag sequences or the express sequence tags (EST), each of which correspond to a particular transcript. EST is short sequences of DNA. These are useful for identifying the gene transcripts. The number of time for which each tag is observed represents the extent of expression of that gene.
The characteristics or the functions of an organism and its body organs are determined by the expression of genes in his genome. So the analysis and the observation of gene expression and the extent of gene expression are helpful to understand the characteristics of the organism. For instance, the isolation of the genes of human pancreas, concatenation and their cloning revealed the pattern characteristics of its functions.
Dr. Victor Velculescuhe developed this SAGE technique in year 1995. This technique is similar to DNA microarrays technique which measures the expression levels of genes quantitatively.
Comparison of SAGE with other technique
Similar to the DNA microarray technique for quantitative measurements of gene expression, but the SAGE technique is based upon sequences of mRNA rather than their hybridization with a radioactive probe which is used in DNA microarrays.
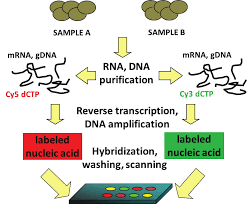
Similarly, the differential display techniques do not provide the comprehensive profiles as SAGE can. So the quantitative measurement of expression levels by SAGE is more precise as it directly counts the number of transcripts. The technique was basically developed to study cancer.
Principle of SAGE Technique
The serial analysis of gene expression technique is based upon the facts that:
- A short sequence tag can have enough of information to identify a particular transcript if it is obtained from that transcript.
- The sequences or the tags can be joined together for sequencing and cloning purposes
- Counting the number of times for which the tags are analyzed provide information about the extent of expression level of the transcript.
Steps Involved In Serial Analysis Of Gene Expression
These are some steps for the SAGE technique:
- Isolation of the mRNA from the tumor cell and then reverse transcriptase activity is applied to create complementary DNA (cDNA). The purpose of this step is that the mRNA is composed of the exons (the coding regions of RNA) only as the interons (the non-coding sequences) are removed before the translation step.
- To synthesize cDNA from mRNA, biotinylation of the primers of mRNA is essential that is covalent attachment of biotin protein to the nucleic acid molecule.
- The cDNA attaches to a supramagnetic particles termed as streptavidin beads and then by the application of restriction endonuclease enzymes they are cleaved at specific restriction sites.
- Sequence of DNA downstream of the restriction site is useless while the upstream sequence is exposed to two oligonucleotide adapters.
Components of adapter sequence
Each of the adapter consists of the following parts:
- Stickey ends that are formed by endonucleases
- A recognition site for the tagging enzyme that is also endonuclease
- A primer specific to each adapter that is later used in PCR reaction
- Adapter is ligated to the cDNA, endonuclease cuts and separates the cDNA from the magnetic beads and the result is that short “tags” of the original cDNA are obtained.
- DNA polymerase activity on these cDNA fragments then creates “blunt ends” of the cDNA.
- These cDNA tag fragments (with adapter primers and AE and Tagging enzyme’s recognition sites attached to it ) are ligated together attaching the two tag sequences together. Also the flanking adapters A and B are attached ate the both ends.These newly constructed fragments termed as “ditags” (two tag sequences) are amplified by the PCR technique using specified primers.
- The ditags fragments are then cleaved and later allowed to be attached with the other ditags. Ligation results in formation of cDNA concatemer/chain with each of the ditag that is separated by a specific recognition site.
- For the replication of these concatemers, these are transformed into the bacterial cell.
- CDNA concatemers are isolated in the next step and then sequenced together by using DNA –sequencers and are quantitatively analyzed by computer.
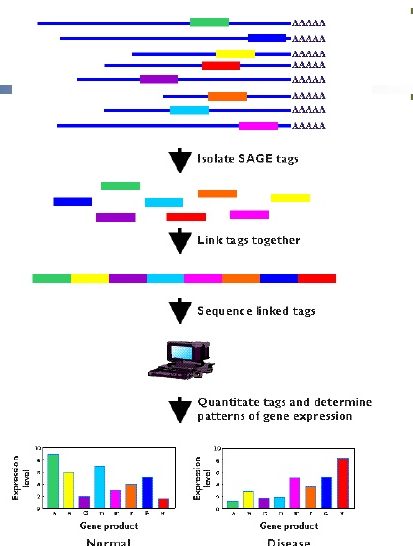
The output of SAGE
- The output of serial analysis of gene expression is in the form of list of short tags. These are sequenced and analyzed.
- A researcher can determine the mRNA origin of tag, which is obtained by SAGE. This is possible by observation of the sequence databases.
- For quantitative analysis, researchers use certain statistical methods to count the tags to determine the extent of expression of genes. For example, the comparison of samples from a normal tissue and from a tumor can be done. This can help to determine in which sample, genes are more or less expressed or active.
Applications of serial analysis of gene expression
SAGE technique is useful for:
- Characterization of the genes expressed in cell under normal as well as in diseased conditions as in cancer
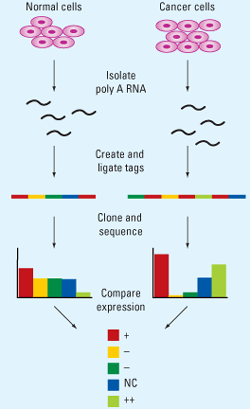
- Analysis of the transcriptosomes- a set of all mRNA molecules of a cell or the cell population.
- Analysis of trancriptosome can be significant for understanding the mechanism and bases of cancer and other cellular differentiation process.
- Comparison of gene expression profiles in normal and the tumor cell
- It provides insight of the P53 gene-mediated apoptosis-the process of programmed cell death
- Analysis of the Adenomatous polyposis gene (APC) that is tumor suppresser gene, to understand its role and gene activation. Also, the factors which affect its expression.
- Use for identification and classification of tumor suppressor gene P53 and its regulation.
- To study the effects of ferredoxin reductase on cancer cells that was dependent on expression of P53 gene.
- Transcriptosome analysis of the oocyte cells help in understanding the cell signaling pathways for its development.
So the serial analysis of gene expression is a useful technique to provide the level of gene expression in cells. These quantitative analysis techniques is valuable tool to understand the expression of genes in various diseases. Moreover, it has its applications for assessment and quality control of drugs and chemicals.